When it comes to parallelism Python has some constraints which have to be taken into consideration before starting coding. I think the biggest one has to do with the Global Interpreter Lock which prevents several threads from executing Python bytecodes at once. Nevertheless you may want to apply concurrency patterns to you code in order to achieve more speed. Besides that you may want to use your cores properly otherwise you’ll end up like this:

In my specific case I wanted to speed up the process of parsing files (as a major features of smalisca). I’ve ended up looking at the muliprocessing package which seemed quite promising:
multiprocessing is a package that supports spawning processes using an API similar to the threading module. The multiprocessing package offers both local and remote concurrency, effectively side-stepping the Global Interpreter Lock by using subprocesses instead of threads. Due to this, the multiprocessing module allows the programmer to fully leverage multiple processors on a given machine. It runs on both Unix and Windows.
In the next steps I’ll try to document my beginners attempts at concurrency in Python.
The idea
Using the -l (location) parameter you can specify the location path where smalisca should
lookup for files before parsing them and afterwards collecting valuable information. Looking closely
at the code I’ve noticed that os.walk already returns a list of “some important information about files and directories.” The multiprocessing documentation refers to the Pool object:
[…] is the Pool object which offers a convenient means of parallelizing the execution of a function across multiple input values, distributing the input data across processes (data parallelism). […]
So I can:
- parallelize the execution of a single function
- use the same input data source for each thread/process
- distribute the input data across processes
Ok, that sounds great!
Some basic code
The basic idea is to “walk” through files and call a function for every single found file. You’d usually start like this:
|
|
But first an input filename list has to be generated:
|
|
In order to achieve concurrency the initial list has to be splitted into several sub-lists
which are then distributed to some “workers” (aka processes). The best way to do this is to
distribute the sub-lists equally to the number of available CPU cores. Given an initial
file_list one could do following:
|
|
But wait a second… What should every process do? Well, the answer is quite obvious: For every file in its sub-list, it has to call a certain function. Afterwards the process has to somehow return the results to the parent process. But how is this done?
Communication between processes
A simple way is to use Queue where you can pass messages back and forth between your processes. In a more complicated example you might use JoinableQueue where you have typical consumer scenario: Processes fetch data, modify it and then push them back into the queue (producer consumer pattern).
In my case a Queue is perfectly fine. That way I have a safe way of letting the process pushing back the results into the queue, before merging all available results globally.
Let’s have a look at some example:
|
|
process_files gets a file list (a sub-list from the big list) and a queue. After
parsing the files, the function will simply put the results into the queue. This
specific type of queue should be thread and process safe as the documentation states.
Collecting results
After all processes are done doing their job, it’s time to collect the results:
|
|
q is the queue used by the processes. Just get all collected results and then you’re
ready to go. You can found the whole code here.
Ok, I’ve shown some introductory example how to use multiprocessing properly by using
a very simplified code. Now let’s move on to a more (or less) complicated scenrio:
Implement parallelism for smalisca’s parsing features.
Smalisca and parallelism
One of the slowest step within smalisca is parsing. And that’s because every directory, every file found within it, has to be parsed sequentially. Even from the beginning of the development I’ve thought concurrency would be a better choice to solve that. Due to the priority of other main functionalities this never happened. Till now :)
As I’m writing this text I also implement this new feature into smalisca. Before I’ve started with the coding part, I had to think about several things:
- Should concurrency be implemented as a core feature of the parsing component?
- Regarding the sub-lists: Should I split file or directory lists?
- How do I collect the results gained from the processes?
- Will there be any major side-effects caused by the concurrency?
Regarding 1: Since I want to keep things simple and clean, I like the idea of a controller which controls the parser. SmaliParser should implement everything that has to do with the parsing itself, but the concurrency should be implemented on-top. That’s why I’ve
decided to parallelize several SmaliParser instances rather than creating processes inside it.
Regarding 2: Due to the fact that the current directory path is used when creating new class objects, I’ve decided to split the directory list, distribute the sub-lists equally across the workers and then collect the results.
Regarding 3: The answer to this one should be quite obvious. One could use a Queue (a thread-safe one like mutiprocessing.Queue) and put the results of every worker
into it. Well that’s pretty much what I’ve done except for the fact that I’ve used proxy objects instead of directly accessed ones. I’ll come to this later one.
Regarding 4: I couldn’t notice any side-effects.
The basic stuff
I didn’t have to write a lot of code. In my case the concurrency has been implemented this way:
- for a given path location, walk the location and return lists of all found directories and files
- split the big list into smaller sub-lists
- every worker/process gets a sub-list
- each worker initiates a
SmaliParserinstance for every directory in its sub-list - after
SmaliParserfinishes its work, the results are pushed into a thread-safeQueue
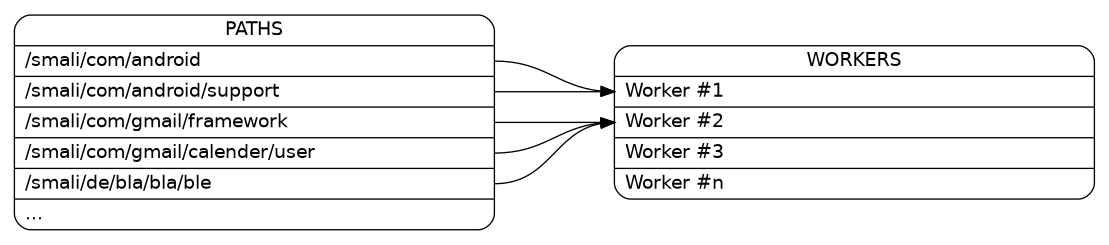
So basically you’ll have a list of directory paths:
Graphviz code
dot {
digraph G {
graph [splines=curve, rankdir = LR, pad=".15", ranksep="1.25", nodesep="2.25"];
node[fontname="FreeSans",fontsize="14",shape=Mrecord,width=7, height=.5];
compound = true;
Bar[label="{\
{PATHS \r |\
<p1>/smali/com/android \l|\
<p2>/smali/com/android/support \l|\
<p3>/smali/com/gmail/framework \l|\
<p4>/smali/com/gmail/calender/user \l|\
<p5>/smali/de/bla/bla/ble \l |\
<pn>... \l \
}\
}", width=8];
}
}

And some workers to do the job:
Graphviz code
dot {
digraph G {
graph [splines=curve, rankdir = LR, pad=".15", ranksep="1.25", nodesep="2.25"];
node[fontname="FreeSans",fontsize="14",shape=Mrecord,width=2, height=.5];
compound = true;
Workers[label="{\
{WORKERS \r |\
<w1>Worker #1 \l|\
<w2>Worker #2 \l|\
<w3>Worker #3 \l|\
<w4>Worker #n \l\
}\
}", width=8];
}
}

In my case the paths have to be distributed to the workers:
Graphviz code
dot {
digraph G {
graph [splines=curve, rankdir = LR, pad=".15", ranksep="1.25", nodesep="2.25"];
node[fontname="FreeSans",fontsize="14",shape=Mrecord,width=2, height=.5];
compound = true;
Bar[label="{\
{PATHS \r |\
<p1>/smali/com/android \l|\
<p2>/smali/com/android/support \l|\
<p3>/smali/com/gmail/framework \l|\
<p4>/smali/com/gmail/calender/user \l|\
<p5>/smali/de/bla/bla/ble \l |\
<pn>... \l \
}\
}", width=5];
Workers[label="{\
{WORKERS \r |\
<w1>Worker #1 \l|\
<w2>Worker #2 \l|\
<w3>Worker #3 \l|\
<w4>Worker #n \l\
}\
}", width=5];
Bar:p1 -> Workers:w1;
Bar:p2 -> Workers:w1;
Bar:p3 -> Workers:w2;
Bar:p4 -> Workers:w2;
Bar:p5 -> Workers:w2;
}
}
So let’s continue with some code examples. A typical parser process would look like this:
|
|
A process will have a list of directories to scan plus a results queue where to put its individual results.
Graphviz code
dot {
digraph G {
graph [splines=curve, rankdir = LR, pad=".15", ranksep="1.25", nodesep="2.25"];
node[fontname="FreeSans",fontsize="14",shape=Mrecord,width=2, height=.5];
compound = true;
Workers[label="{\
{WORKERS \r |\
<w1>Worker #1 \l|\
<w2>Worker #2 \l|\
<w3>Worker #3 \l|\
<w4>Worker #n \l\
}\
}", width=4];
Queue [label="Results Queue", shape=box3d, width=4];
Workers:w1 -> Queue;
Workers:w2 -> Queue;
Workers:w3 -> Queue;
Workers:w4 -> Queue;
}
}
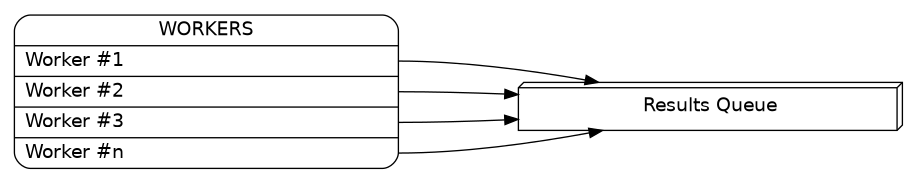
A controller should create and create the workers. Afterwards it should collect the results:
|
|
Pretty straightforward, isn’t it? Split the lists into sub-lists, assign each worker a sub-list and let them run. Then join the processes and collect the results.
Concurrency caveats
In some initial code I’ve had, I was using a simple multiprocessing.Queue() to collect
the results. After some testing I’ve noticed that my processes never terminated.
Sometimes (in fact always) it’s a good idea to take a closer look at the documentation.
Looking at the programming guidelines I’ve read following:
Joining processes that use queues
Bear in mind that a process that has put items in a queue will wait before terminating until all the buffered items are fed by the “feeder” thread to the underlying pipe. (The child process can call the Queue.cancel_join_thread() method of the queue to avoid this behaviour.)
Ä This means that whenever you use a queue you need to make sure that all items which have been put on the queue will eventually be removed before the process is joined. Otherwise you cannot be sure that processes which have put items on the queue will terminate. Remember also that non-daemonic processes will be automatically be joined.
So my previous implementation was prone to a deadlock:
|
|
OK, leason lerned! And then I’ve found this post which deals with multiprocessing.Managers. In my current code I’m also using a Queue but proxied:
|
|
In this case result_queue is also a Queue wrapped by a proxy. As stated here:
A proxy is an object which refers to a shared object which lives (presumably) in a different process. The shared object is said to be the referent of the proxy. Multiple proxy objects may have the same referent. .. An important feature of proxy objects is that they are picklable so they can be passed between processes. Note, however, that if a proxy is sent to the corresponding manager’s process then unpickling it will produce the referent itself.
Ahhhh! Now that we’ve implemented it (hopefully) the clean way, let’s have a look at some metrics.
Metrics
Of course I wanted to somehow measure my improvements. Having that said don’t expect huge improvements. It kind of surprised me too. I’ve compared the execution time (just for the parsing job) between
- old and new version of smalisca
- using different number of jobs/workers
I’ve used my Laptop (Dell XPS 13 with Intel Core i7) for the testings. Using a larger code repository (bigger than the FakeBanker one) I’ve first ran the old smalisca version (0.1). This what I’ve got:
|
|
As you see it takes ca. 3.5 seconds to finish the parsing. OK, now what about the concurrent version of smalisca?
|
|
5.5 seconds!!! It takes longer! Obviously the Queue processing and starting new processes increases
execution time. Ok, now let’s try with more jobs (=8):
|
|
Pretty much the same results. And here is the overall results table:
| Run | Non-Concurrent (v0.1) | Concurrent (v0.1-dev) | Using Jobs = 8 |
|---|---|---|---|
| 1 | 3.61 | 5.52 | 5.66 |
| 2 | 3.66 | 5.50 | 5.80 |
| 3 | 3.66 | 5.59 | 5.57 |
| 4 | 3.64 | 5.48 | 5.49 |
| 5 | 3.67 | 5.42 | 5.76 |
| 6 | 3.56 | 5.79 | 5.62 |
| 7 | 3.55 | 5.50 | 5.54 |
| 8 | 3.63 | 5.39 | 5.63 |
| 9 | 3.52 | 5.38 | 5.73 |
| 10 | 3.55 | 5.44 | 5.43 |
Lessons learned
- Concurrency is NOT easy
- It may not always have a big (positive) impact on the performance of your code
- There are a lot of side-effects (related to concurrency in general) which you’ll have to pay attention to
- I need a really big code repository to test against in order to make sure I’ve implemented it correctly
- Despite all those kind of problems, it’s fun! :)
- Have a look at the commit details in the develop branch.